PROTOCOL
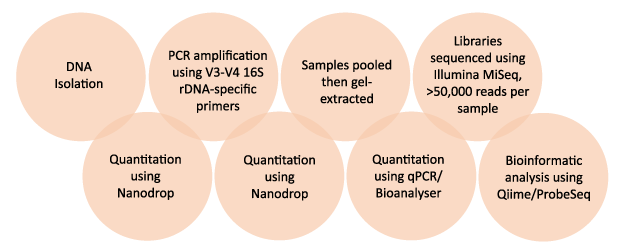
A modification of the protocol as described by Caporaso et. al (2011) was used.
- 10-50 ng of DNA were PCR-amplified using V3-V4 primers and 5 PrimeHotMaster Mix.
- PCR samples were purified using AMPure beads.
- 100 ng of each library was pooled, gel-purified, and quantified using a bioanalyser and subsequently with qPCR.
- 12 pM of the library mixture, spiked with 20% Phix, was run on the MiSeq.
The V3-V4 primers was used as follows:
~341F (forward primer)
AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTCCTACGGGAGGCAGCAG
~806R (reverse primer)
CAAGCAGAAGACGGCATACGAGATNNNNNNNNNNNNAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT
Sequences of the primers are in bolded italics
12 N’s (in red) designate barcode sequences
These primers are slightly different from those primers used by Illumina.
